



|
|
Contact Sarah Elgin at
selgin@biology.wustl.edu
or (314) 935-5348. | |||
|
|
Lab Telephone: (314) 935-6837 |
FAX: (314) 935-5125 | ||
|
|
US Mail: Washington UniversityDept. Biology - CB 1229 One Brookings Drive St. Louis, MO 63130. |
|
FedEx etc: Washington University / Biology133 Rebstock Forsyth at Tolman Way St. Louis, MO 63130. |
|
We are interested in the role
that chromatin structure plays in gene regulation, considering both
effects from packaging large domains and local effects of the
nucleosome array. We work with Drosophila, combining
biochemical, genetic and cytological approaches. We have used a
transposable P-element containing a copy of the white gene, a visible marker for
gene silencing, and a copy of hsp26, a well-characterized
inducible gene, to examine the effect of insertion into different
chromosomal domains. While these genes are fully active in
euchromatic domains, silencing similar to Position Effect Variegation
can be observed when the P-element is inserted into pericentric
heterochromatin, telomeres, or the small fourth chromosome (see
figure below). Further investigations to examine the mechanism(s) of
gene silencing are in progress; changes in the local nucleosome
array, as well as spatial organization in the nucleus, appear
critical.
Earlier work in the lab identified Heterochromatin
Protein 1 (HP1) as a protein preferentially associated with the
pericentric heterochromatin, and in a banded pattern with the small
fourth chromatin. Subsequent analysis showed that HP1 is encoded by
Su(var)2-5; both
mutations that would be expected to reduce the level of HP1, and a
point mutation, result in suppression of Position Effect Variegation,
suggesting that HP1 plays a role in establishing heterochromatic
structure. All of the known mutant alleles of Su(var)2-5 result in greater
expression from our test transgenes inserted in pericentric
heterochromatin and the fourth chromosome, but have no impact on
expression of the same transgenes in the telomeres of the second or
third chromosome, indicating a difference in these heterochromatic
domains. Work is ongoing to identify additional
heterochromatin-associated proteins, screening for interactions with
HP1. A candidate has been identified using a yeast two-hybrid screen;
this protein, HP2, shows a similar distribution pattern on polytene
chromosomes.
Transgene inserts showing a variegating phenotype
have been recovered not only in the pericentric heterochromatin and
telomere of the fourth chromosome, but within the banded region of
that chromosome. Recently we have identified several P-element
insertion sites on the fourth chromosome that allow full gene
expression, suggesting that this region has closely interspersed
heterochromatic and euchromatic domains. Utilizing such lines, we
plan to create a functional map, allowing characterization of such
domains and their boundaries. Results to date indicate consistent
differences in the chromatin structure of heterochromatic and
euchromatic domains.
We are also looking at the effect of a homeotic
regulatory signal, PRE, on hsp26 structure and function
(collaboration with V. Pirrotta, Geneva); such signals may maintain
silencing by altering chromatin structure. Thus, shifts in the
nucleosome array, and concommitant shifts in higher order structure,
might be a key distinction between activatable and silent genes
programmed during development.
The above studies require a detailed knowledge of
the test gene, hsp26. Previous work from our lab and others has shown that
correct assembly of the hsp26 regulatory region in an
activatable form requires two (CT)n
sites, which bind GAGA factor. We are examining the effect of various
mutations in the hsp26 promoter on GAGA factor function and chromatin assembly, in
particular asking whether the presence of an immediately adjacent
TFIID binding site is critical (collaboration with D. Gilmour, Penn
State).
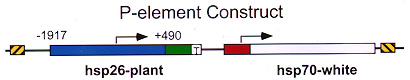
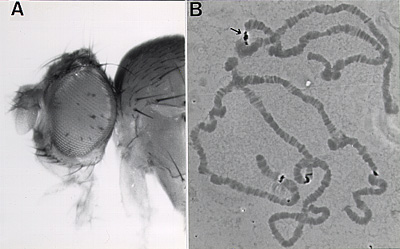
|
Our P-element construct (based on vector A412 from V. Pirrotta) contains a visible marker for variegation, hsp70-white, and a marked copy of hsp26 for subsequent studies of chromatin structure. Several fly lines have been recovered showing a PEV phenotype (A); all have P element inserts in the pericentric heterochromatin (as shown for this case by in situ hybridization to the polytene chromosomes with the entire P element; see B), telomeres, or the fourth chromosome. (Note that the P element also hybridizes to the sites of the endogenous white, hsp70, and hsp26 genes.) See Elgin et al, 1993, and Wallrath and Elgin, 1995; full citations and abstracts below. |