Chromatin: A Practical Approach. 1998, Oxford University Press: New York, Tokyo.
Department of Biology, Washington University, St. Louis, Missouri 63130, USA.
This chapter presents some of the techniques we have developed for the analysis of chromatin structure using a variety of nucleases to discern patterns of accessibility along the chromatin fiber. It includes mapping with DNase I, micrococcal nuclease, and restriction enzymes using embryos, larvae or adult flies as the starting material.
Mol Cell Biol 1993 May;13(5):2802-2814
Department of Biology, Washington University, St. Louis, Missouri 63130.
Previous analysis of the hsp26 gene of Drosophila melanogaster has shown that in addition to the TATA box and the proximal and distal heat shock elements (HSEs) (centered at -59 and -340, relative to the start site of transcription), a segment of (CT)n repeats at -135 to -85 is required for full heat shock inducibility (R.L. Glaser, G.H. Thomas, E.S. Siegfried, S.C.R. Elgin, and J.T. Lis, J. Mol. Biol. 211:751-761, 1990). This (CT)n element appears to contribute to formation of the wild-type chromatin structure of hsp26, an organized nucleosome array that leaves the HSEs in nucleosome-free, DNase I-hypersensitive (DH) sites (Q. Lu, L.L. Wallrath, B.D. Allan, R.L. Glaser, J.T. Lis, and S.C.R. Elgin, J. Mol. Biol. 225:985-998, 1992). Inspection of the sequences upstream of hsp26 has revealed an additional (CT)n element at -347 to -341, adjacent to the distal HSE. We have analyzed the contribution of this distal (CT)n element (-347 to -341), the proximal (CT)n element (-135 to -85), and the two HSEs both to the formation of the chromatin structure and to heat shock inducibility. hsp26 constructs containing site-directed mutations, deletions, substitutions, or rearrangements of these sequence elements have been fused in frame to the Escherichia coli lacZ gene and reintroduced into the D. melanogaster genome by P-element-mediated germ line transformation. Chromatin structure of the transgenes was analyzed (prior to gene activation) by DNase I or restriction enzyme treatment of isolated nuclei, and heat-inducible expression was monitored by measuring beta-galactosidase activity. The results indicate that mutations, deletions, or substitutions of either the distal or the proximal (CT)n element affect the chromatin structure and heat-inducible expression of the transgenes. These (CT)n repeats are associated with a nonhistone protein(s) in vivo and are bound by a purified Drosophila protein, the GAGA factor, in vitro. In contrast, the HSEs are required for heat-inducible expression but play only a minor role in establishing the chromatin structure of the transgenes. Previous analysis indicates that prior to heat shock, these HSEs appear to be free of protein. Our results suggest that GAGA factor, an abundant protein factor required for normal expression of many Drosophila genes, and heat shock factor, a specific transcription factor activated upon heat shock, play distinct roles in gene regulation: the GAGA factor establishes and/or maintains the DH sites prior to heat shock induction, while the activated heat shock factor recognizes and binds HSEs located within the DH sites to trigger transcription.
|
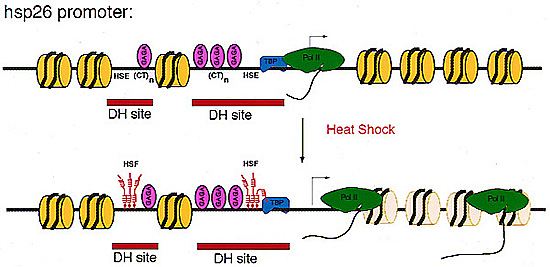
Pre-heat shock chromatin structure of hsp26 transgenes. The D.melanogaster hsp26 gene is assembled into a precise chromatin structure prior to heat shock. A specifically positioned nucleosome (yellow disk) lies between two nucleosome-free DNase I hypersensitive sites (DH sites; red bars). These DH sites encompass the heat shock elements (HSEs) and (CT)n elements which bind GAGA factor. GAGA bound to high affinity sites is colored mauve. The TATA binding protein (TBP; blue) is present at the promoter prior to heat shock, and RNA polymerase II (green) has paused after synthesizing a short transcript (see Lis and Wu, 1993). After heat shock, heat shock transcription factor binds to the HSEs and triggers transcription; there is a perturbation of the downstream nucleosome array during transcription. |
EMBO J 1995 Oct 2;14(19):4738-4746
Department of Biology, Washington University, St Louis, MO 63130, USA.
The regulatory region of Drosophila melanogaster hsp26 includes a positioned nucleosome located between the two DNase I hypersensitive (DH) sites that encompass the critical heat shock elements (HSEs). To test the role of this nucleosome in regulated expression, transgenic flies containing hsp26-lacZ fusion genes with alterations in the nucleosome-associated region have been generated. The positioned nucleosome is associated with a DNA sequence that does not itself contain any critical regulatory elements for heat shock-inducible expression. The nucleosome-associated sequence can be deleted, reversed, duplicated or replaced by a random sequence with no significant effect on DH site formation and gene expression. Analyses of hsp26 and hsp70 transgenes with spacing changes within the promoter region indicate that the location of the (CT)n.(GA)n elements dictates the location of DH site formation. Wrapping the DNA between the regulatory elements around a nucleosome is as effective for gene expression as placing the regulatory elements close to each other. A loss of inducible gene expression was observed when the nucleosome-associated DNA was replaced with sequences which appear to misdirect nucleosome placement. The results indicate considerable flexibility in the spacing between DH regulatory sites.
Cold Spring Harb Symp Quant Biol 1993;58:83-96
Department of Biology, Washington University, St. Louis, Missouri 63130.
A review of the various experiments (nuclease digestion studies, examination of the effect of various mutations in hsp26 transgenes, etc) that have led to the detailed model of the chromatin structure of the hsp26 gene.
Curr Biol 1995 Mar 1;5(3):238-241
Department of Biology, Washington University, St Louis, Missouri 63130, USA.
Recent results suggest that the Drosophila transcriptional activator known as GAGA factor functions by influencing chromatin structure. This minireview tabulates some of the genes regulated by GAGA factor, and some of the GAGA factor binding sites, while summarizing the evidence that GAGA factor acts primarily at the chromatin level. GAGA factor has been shown to be a sequence-specific DNA binding protein. It is encoded by Trl; mutations in this gene result in an enhancement of Position Effect Variegation (see Farkas et al, 1994, Nature 371:806-808). Thus GAGA factor may help to determine the local nucleosome array, and by so doing may influence higher order chromatin structure.
|
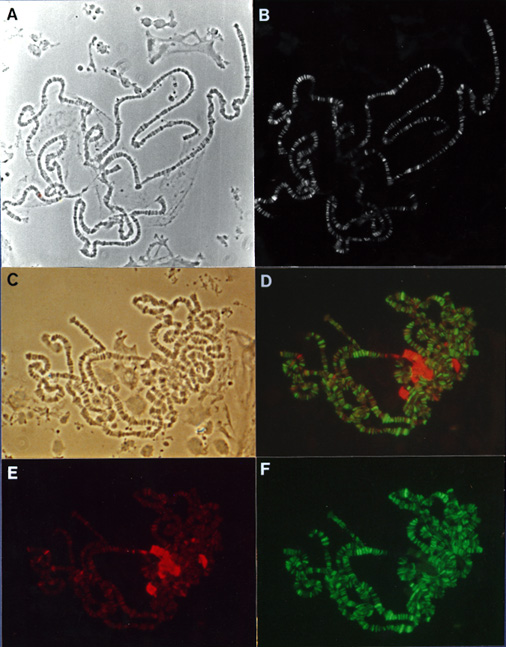
Indirect immunofluorescence of
D. melanogaster polytene chromosomes stained with antibodies
against the GAGA factor and HP1 reveals predominantly
exclusive patterns of protein distribution. |
Nucleic Acids Res 1997 Aug 15;25(16):3345-3353
Department of Biology, University of Rochester, Rochester, NY 14627, USA. tip@cb.biology.rochester.edu
The GAGA transcription factor of Drosophila melanogaster is ubiquitous and plays multiple roles. Characterization of cDNA clones and detection by domain- specific antibodies has revealed that the 70-90 kDa major GAGA species are encoded by two open reading frames producing GAGA factor proteins of 519 amino acids (GAGA-519) and 581 amino acids (GAGA-581), which share a common N-terminal region that is linked to two different glutamine-rich C-termini. Purified recombinant GAGA-519 and GAGA-581 proteins can form homomeric complexes that bind specifically to a single GAGA sequence in vitro. The two GAGA isoforms also function similarly in transient transactivation assays in tissue culture cells and in chromatin remodeling experiments in vitro . Only GAGA-519 protein accumulates during the first 6 h of embryogenesis. Thereafter, both GAGA proteins are present in nearly equal amounts throughout development; in larval salivary gland nuclei they colocalize completely to specific regions along the euchromatic arms of the polytene chromosomes. Coimmunoprecipitation of GAGA-519 and GAGA-581 from crude nuclear extracts and from mixtures of purified recombinant proteins, indicates direct interactions. We suggest that homomeric complexes of GAGA-519 may function during early embryogenesis; both homomeric and heteromeric complexes of GAGA-519 and GAGA-581 may function later.
|
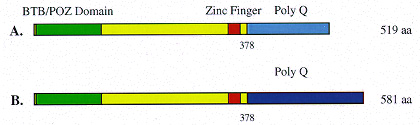
GAGA factor isoforms. Two classes of cDNAs have been recovered for the GAGA factor to date (Soeller et al, 1993; Benyajati et al, paper above). The predicted proteins both contain a single C2-H2 type Zn-finger. The amino-terminus - the BTB or POZ domain - shares homology to a large number of eukaryotic and viral proteins and may serve as a protein-protein interaction domain (Zollman et al, 1994; Bardwell and Treisman, 1994). These GAGA factor isoforms diverge at amino acid 378; both proteins contain a glutamine-rich carboxy terminus. |
Mol Biol Cell 1992 Jun;3(6):593-602
Department of Biology, Washington University, St. Louis, Missouri 63130.
H2AvD, a Drosophila melanogaster histone variant of the H2A.Z class, is encoded by a single copy gene in the 97CD region of the polytene chromosomes. Northern analysis shows that the transcript is expressed in adult females and is abundant throughout the first 12 h of embryogenesis but then decreases. The H2AvD protein is present at essentially constant levels in all developmental stages. Using D. melanogaster stocks with deletions in the 97CD region, we have localized the H2AvD gene to the 97D1-9 interval. A lethal mutation in this interval, l(3)810, exhibits a 311-base pair deletion in the H2AvD gene, which removes the second exon. P-element mediated transformation using a 4.1-kilobase fragment containing the H2AvD gene rescues the lethal phenotype. H2AvD is therefore both essential and continuously present, suggesting a requirement for its utilization, either to provide an alternative capability for nucleosome assembly or to generate an alternative nucleosome structure.